Data Science Analysis of Running Form
Analyzing the Correlation Between Running Stride Length and Average Pace Per Mile
from datascience import *
%matplotlib inline
import matplotlib.pyplot as plots
plots.style.use('fivethirtyeight')
import numpy as np
Importing the Data
The data generating process was as follows: I went on my run. Once I finished my run, I saved it. I uploaded the run to my iPhone via bluetooth. Once the running data uploaded, I exported a CSV file of the data to my computer.
Here is a link to the google sheets version of the data. For reference, the watch I used to collect this data is the COROS Pace 2.
As a side note, this statistical analysis only takes into account one run. In the future I plan to run a more comprehensive analysis because it is essential to have a variety of data to truly prove a statistical relationship. However, this is a good jumping-off point.
The following running data is from an 18 mile long run. I started at 6:39 mile pace and closed in 5:27 for the last mile. As it was progressive, I chose to analyze the linear relationship between stride length as running pace increases. For the actual analysis, I used the column that contains minutes per kilometer rather than minutes per mile. We are also, for the purpose of this project, holding constant external factors such as elevation gain per mile, heart rate, and prior muscle fatigue.
progressive_longrun_data = Table.read_table("ProgressiveLongRun.csv")
progressive_cleaned = progressive_longrun_data.take(np.arange(0,18,1))
progressive_cleaned.show()
| Split | Time | Moving Time | Distance | Elevation Gain | Elev Loss | Avg Pace | Avg Moving Pace | Best Pace | Avg Run Cadence | Max Run Cadence | Avg Stride Length | Avg HR | Max HR | Avg Temperature | Calories |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0:06:39 | 0:06:39 | 1.61 | 0 | 31 | 0:04:08 | 0:04:05 | 0:03:39 | 82 | 84 | 148 | 128 | 147 | 24 | 48 |
| 2 | 0:06:33 | 0:06:33 | 1.61 | 6 | 30 | 0:04:04 | 0:04:03 | 0:03:46 | 82 | 83 | 150 | 141 | 145 | 22 | 63 |
| 3 | 0:06:33 | 0:06:33 | 1.61 | 27 | 21 | 0:04:04 | 0:04:08 | 0:03:47 | 83 | 86 | 149 | 149 | 154 | 20 | 59 |
| 4 | 0:06:31 | 0:06:31 | 1.61 | 16 | 31 | 0:04:03 | 0:04:04 | 0:03:54 | 82 | 85 | 150 | 151 | 153 | 21 | 70 |
| 5 | 0:06:33 | 0:06:33 | 1.61 | 56 | 15 | 0:04:04 | 0:04:06 | 0:03:14 | 84 | 89 | 147 | 148 | 156 | 21 | 58 |
| 6 | 0:06:31 | 0:06:31 | 1.61 | 5 | 31 | 0:04:03 | 0:04:02 | 0:03:52 | 82 | 85 | 150 | 155 | 165 | 23 | 72 |
| 7 | 0:06:29 | 0:06:29 | 1.61 | 49 | 0 | 0:04:02 | 0:04:01 | 0:03:51 | 84 | 87 | 148 | 167 | 172 | 23 | 70 |
| 8 | 0:06:31 | 0:06:27 | 1.61 | 34 | 4 | 0:04:00 | 0:04:01 | 0:03:47 | 84 | 86 | 148 | 165 | 173 | 24 | 68 |
| 9 | 0:06:27 | 0:06:27 | 1.61 | 7 | 29 | 0:04:00 | 0:04:01 | 0:03:48 | 83 | 85 | 151 | 159 | 167 | 25 | 65 |
| 10 | 0:06:21 | 0:06:21 | 1.61 | 0 | 34 | 0:03:57 | 0:03:57 | 0:03:52 | 83 | 84 | 154 | 162 | 170 | 25 | 78 |
| 11 | 0:06:15 | 0:06:15 | 1.61 | 0 | 35 | 0:03:53 | 0:03:53 | 0:03:38 | 83 | 85 | 155 | 165 | 169 | 25 | 69 |
| 12 | 0:06:12 | 0:06:12 | 1.61 | 34 | 24 | 0:03:51 | 0:03:53 | 0:03:37 | 84 | 88 | 154 | 165 | 170 | 24 | 68 |
| 13 | 0:06:05 | 0:06:05 | 1.61 | 15 | 32 | 0:03:47 | 0:03:49 | 0:03:28 | 84 | 89 | 157 | 159 | 167 | 24 | 66 |
| 14 | 0:05:55 | 0:05:55 | 1.61 | 46 | 17 | 0:03:41 | 0:03:41 | 0:02:46 | 86 | 90 | 159 | 164 | 176 | 23 | 67 |
| 15 | 0:05:51 | 0:05:51 | 1.61 | 17 | 30 | 0:03:38 | 0:03:39 | 0:03:23 | 84 | 86 | 163 | 168 | 173 | 24 | 71 |
| 16 | 0:05:46 | 0:05:46 | 1.61 | 48 | 1 | 0:03:35 | 0:03:38 | 0:03:29 | 87 | 89 | 162 | 171 | 177 | 23 | 72 |
| 17 | 0:05:39 | 0:05:39 | 1.61 | 38 | 5 | 0:03:31 | 0:03:32 | 0:03:24 | 86 | 89 | 165 | 172 | 179 | 23 | 61 |
| 18 | 0:05:27 | 0:05:27 | 1.61 | 9 | 22 | 0:03:23 | 0:03:21 | 0:02:53 | 87 | 91 | 169 | 173 | 180 | 24 | 74 |
Initial Visualization of the Data
We want to look at an initial visualization of the data so that we can see if there are any obvious patterns. Upon initial inspection, there looks to be good possibility for a linear relationship between average running pace and average stride length.
progressive_cleaned.scatter("Avg Pace", "Avg Stride Length")
plots.xlabel("Average Running Pace (Minutes per Kilometer)")
plots.ylabel("Average Stride Length (Centimeters)")
Text(0, 0.5, 'Average Stride Length (Centimeters)')
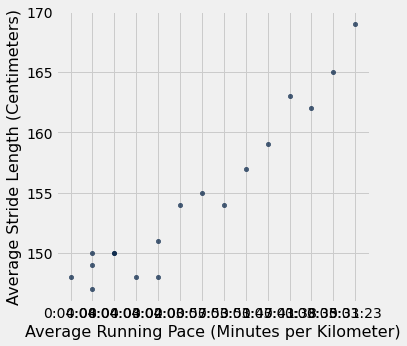
In the initial visualization of the data, it becomes apparent that the minutes and second display on the x axis isn’t very elegant. Consequently, I spent the following lines of code converting the average pace per kilometer from a minutes and seconds version to purely seconds form.
import datetime
string_version = [str(i) for i in progressive_cleaned.column("Avg Pace")]
#modifying the running time in minutes per kilometer from elements in a numpy array to individual strings
updated_list = []
#creating an empty list that we can append the datetime version of the strings to
for i in string_version:
date_time_version = datetime.datetime.strptime(i, "%H:%M:%S")
updated_list.append(date_time_version)
updated_list
final_list = []
#the final list for which we are going to append the time in seconds per kilometer
for i in updated_list:
a_timedelta = i - datetime.datetime(1900, 1, 1)
seconds = a_timedelta.total_seconds()
final_list.append(int(seconds))
running_pace_vs_stride_length = Table().with_column("Average Running Pace", final_list).with_column("Average Stride Length", progressive_cleaned.column("Avg Stride Length"))
running_pace_vs_stride_length.show()
| Average Running Pace | Average Stride Length |
|---|---|
| 248 | 148 |
| 244 | 150 |
| 244 | 149 |
| 243 | 150 |
| 244 | 147 |
| 243 | 150 |
| 242 | 148 |
| 240 | 148 |
| 240 | 151 |
| 237 | 154 |
| 233 | 155 |
| 231 | 154 |
| 227 | 157 |
| 221 | 159 |
| 218 | 163 |
| 215 | 162 |
| 211 | 165 |
| 203 | 169 |
running_pace_vs_stride_length.scatter("Average Running Pace", "Average Stride Length")
plots.xlabel("Average Running Pace (Seconds per Kilometer)")
plots.ylabel("Average Stride Length (Centimeters)")
Text(0, 0.5, 'Average Stride Length (Centimeters)')
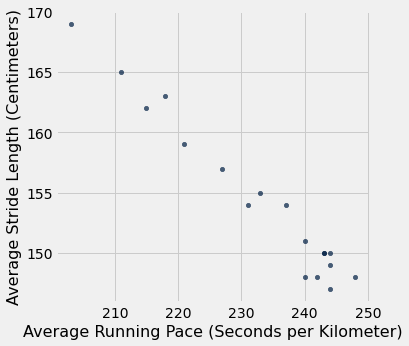
Now we have cleaned the data, dropped the unnecessary parts, and have a simple visualization of what we are going to look at in further detail. We have a two column table containing what we would like to analyze for this particular moment. Time for some statistical testing.
# Creating some functions that we will use to analyze the data.
#We need standard units so that we can run our correlation function.
def standard_units(x):
""" Converts an array x to standard units """
return (x - np.mean(x)) / np.std(x)
def correlation(t, x, y):
""" Computes correlation: t is a table, and x and y are column names """
x_su = standard_units(t.column(x))
y_su = standard_units(t.column(y))
return np.mean(x_su * y_su)
#generating the correlation coefficient
correlation(running_pace_vs_stride_length, "Average Running Pace", "Average Stride Length")
-0.9819409345425044
Wow! We generated a correlation coefficient of -0.98, which means there is almost an exactly proportionate decrease in stride length for an increase in the amount of seconds it take to complete each kilometer! However, a correlation coefficient alone is not enough. We need to also look at a linear regression line and plot the residuals.
#our slope and intercept functions
def slope(t, x, y):
""" Computes the slope of the regression line, like correlation above """
r = correlation(t, x, y)
y_sd = np.std(t.column(y))
x_sd = np.std(t.column(x))
return r * y_sd / x_sd
def intercept(t, x, y):
""" Computes the intercept of the regression line, like slope above """
x_mean = np.mean(t.column(x))
y_mean = np.mean(t.column(y))
return y_mean - slope(t, x, y)*x_mean
slope_for_regression_line = slope(running_pace_vs_stride_length, "Average Running Pace", "Average Stride Length")
slope_for_regression_line
-0.48736086175942556
intercept_for_regression_line = intercept(running_pace_vs_stride_length, "Average Running Pace", "Average Stride Length")
intercept_for_regression_line
267.67321364452425
#generating the predicted stride length from running pace and adding that to the table
predictions = slope_for_regression_line * running_pace_vs_stride_length.column("Average Running Pace") + intercept_for_regression_line
predictions
array([146.80771993, 148.75716338, 148.75716338, 149.24452424,
148.75716338, 149.24452424, 149.7318851 , 150.70660682,
150.70660682, 152.16868941, 154.11813285, 155.09285458,
157.04229803, 159.9664632 , 161.42854578, 162.89062837,
164.84007181, 168.73895871])
run_with_predictions = running_pace_vs_stride_length.with_column("Predicted Stride Length", predictions)
run_with_errors = run_with_predictions.with_column("Error", run_with_predictions.column("Average Stride Length") - run_with_predictions.column("Predicted Stride Length"))
run_with_errors.show()
| Average Running Pace | Average Stride Length | Predicted Stride Length | Error |
|---|---|---|---|
| 248 | 148 | 146.808 | 1.19228 |
| 244 | 150 | 148.757 | 1.24284 |
| 244 | 149 | 148.757 | 0.242837 |
| 243 | 150 | 149.245 | 0.755476 |
| 244 | 147 | 148.757 | -1.75716 |
| 243 | 150 | 149.245 | 0.755476 |
| 242 | 148 | 149.732 | -1.73189 |
| 240 | 148 | 150.707 | -2.70661 |
| 240 | 151 | 150.707 | 0.293393 |
| 237 | 154 | 152.169 | 1.83131 |
| 233 | 155 | 154.118 | 0.881867 |
| 231 | 154 | 155.093 | -1.09285 |
| 227 | 157 | 157.042 | -0.042298 |
| 221 | 159 | 159.966 | -0.966463 |
| 218 | 163 | 161.429 | 1.57145 |
| 215 | 162 | 162.891 | -0.890628 |
| 211 | 165 | 164.84 | 0.159928 |
| 203 | 169 | 168.739 | 0.261041 |
root_mean_squared_error = np.mean(run_with_errors.column("Error") ** 2) ** 0.5
root_mean_squared_error
1.2311567965332846
run_with_errors.scatter("Average Running Pace")
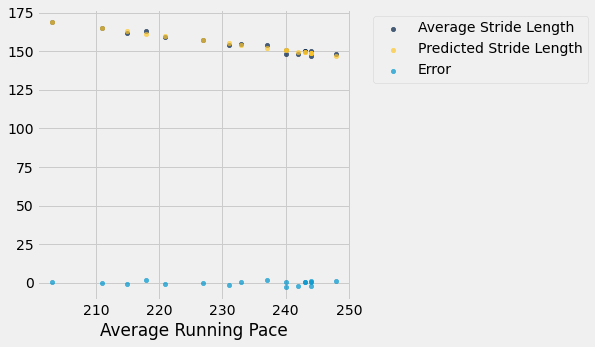
Taking A Closer Look at the Residuals
From that plot, we can tell that the trend in the residuals looks good. The aim of any linear regression estimate is to generate residuals that appear as white noise. However, the plot is a little bit zoomed out. Let’s manipulate the x and y axis to generate a closer look at the residuals.
run_with_errors.scatter("Average Running Pace", "Error")
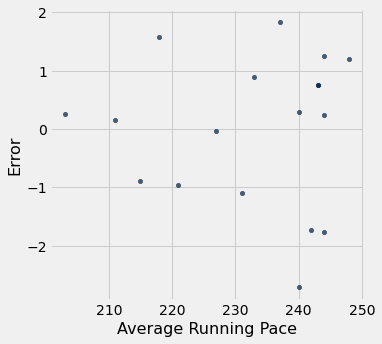
mean_stride_length = np.mean(run_with_errors.column("Average Stride Length"))
mean_stride_length
154.38888888888889
maximum_error = max(abs(run_with_errors.column("Error")))
maximum_error
2.706606822262131
2 / 154.38
0.012955045990413267
root_mean_squared_error
1.2311567965332846
root_mean_squared_error / mean_stride_length
0.007974387311118792
With this zoomed in look, we can tell that most of our errors hover around the range +- 2. This means that our usual error is around 2 centimeters. That’s not a bad error. Given the mean stride length of 154, the predicted percentage of stride length that we will be off by is about 1.3 percent. That’s a darn good estimate. If we take into consideration the RMSE of 1.23, we take 1.23 / 154.38 and get a result of 0.007, which is 0.7 percent.
Conclusion
From analysis of the errors, and correlation coefficient of -0.98, we can conclude that linear prediction is a good indicator for calculating my stride length, given a running pace between 200 and 300 seconds per kilometer.
